About us
We are an interdisciplinary group using archaeology, biomolecular science, and computational genomics to reconstruct health, disease, and population history from the Neolithic to the early modern period. Our work spans pathogen genomics, ancient human population dynamics, and the development of new substrates and methods for biomolecular analysis.
We are best known for research that traces the evolution and impact of infectious diseases (including Yersinia pestis, Mycobacterium leprae, HSV-1, and Haemophilus influenzae), integrates cemetery- and community-scale perspectives on past epidemics, and refines how we interpret mortuary practices and sex/gender bias in the archaeological record. In parallel, we advance ancient biomolecule recovery by validating unconventional reservoirs—such as dental dentin metabolites, bone-adhered sediments, and mineral concretions—that preserve DNA, proteins, and metabolites. Methodologically, we contribute pipelines for ultra-low-coverage genomes and targeted capture, enabling robust inference from challenging material.
Geographically our projects focus on Britain and Europe—with case studies from Cambridgeshire before and after the Black Death, Roman and medieval Britain, and the Italian Peninsula—while also engaging with broader Eurasian and North American histories. Themes include population turnover and continuity (e.g., Steppe-related ancestry shifts; limited Roman genetic impact in rural Britain), community responses to crises (from medieval Cambridge to 17th-century Scotland), and the long-term ecology of pathogens.
We value open, reproducible science and ethical, collaborative practice with museums, communities, and descendant groups. Our lab trains researchers across wet-lab, proteomic/metabolomic, and computational skills, and we share protocols, code, and curated datasets to help the field move forward.
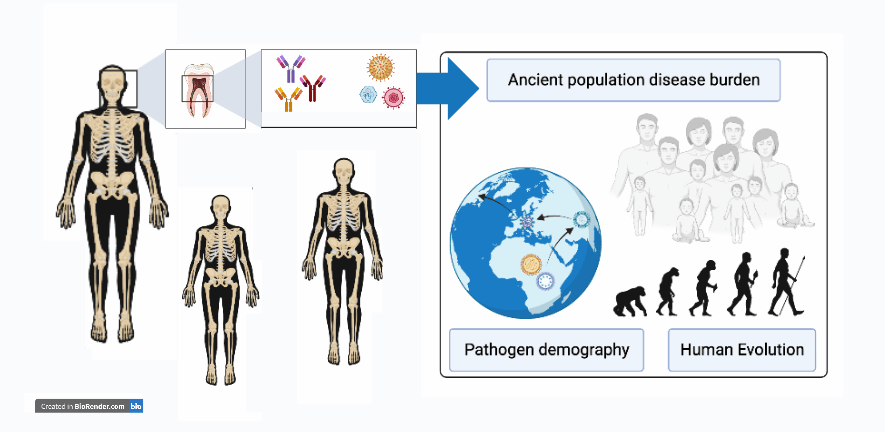
Current research projects
Ancient Antibodies
Identifying pathogen exposure via immunological memory in ancient populations using proteins and DNA: Developing new methods to identify pathogen exposure in ancient populations by analyzing the immunological memory encoded in preserved antibodies and DNA within human skeletal remains. This research aims to provide a deeper understanding of human-pathogen interactions and the evolution of the human immune system by establishing a new field called "Ancient Immunomics".
Palaeo proteomics
Developing methods to isolate and sequence ancient pathogens and immune-response proteins from skeletal tissues.
Ancient DNA (aDNA)
Identifying ancient viruses and pathogens from skeletal remains using aDNA techniques.
Better understanding the sources of DNA in skeletal remains.
Key publications
Badillo-Sanchez, D. A., Jones, D. J. L., Guellil, M., Inskip, S. A., & Scheib, C. L. (2023). Human archaeological dentin as source of polar and less polar metabolites for untargeted metabolomic research: The case of Yersinia pestis. Metabolites, 13(5). https://doi.org/10.3390/metabo13050588
Bonucci, B., de-Dios, T., Barbieri, R., Thompson, J. E., Panella, S., Radina, F., Sivilli, S., Kabral, H., Solnik, A., Tafuri, M. A., Robb, J., & Scheib, C. L. (2025). Ancient microbial DNA and proteins preserve in concretions covering human remains. iScience, 113182, 113182.
Cessford, C., Scheib, C. L., Guellil, M., Keller, M., Alexander, C., Inskip, S. A., & Robb, J. E. (2021). Beyond Plague Pits: Using Genetics to Identify Responses to Plague in Medieval Cambridgeshire. European Journal of Archaeology, 1–23.
de-Dios, T., Bonucci, B., Barbieri, R., Kushniarevich, A., D’Atanasio, E., Dittmar, J. M., Cessford, C., Solnik, A., Robb, J. E., Warinner, C., Oras, E., & Scheib, C. L. (2025). Bone adhered sediments as a source of target and environmental DNA and proteins. Molecular Biology and Evolution, msaf202, msaf202.
Dittmar, J., Crozier, R., de-Dios, T., Scheib, C. L., Armstrong, J. W., Pape, J., MacLennan, R., Craig, R., & Oxenham, M. (2024). The final plague outbreak in Scotland 1644-1649: Historical, archaeological, and genetic evidence. PloS One, 19(11), e0306432.
Elliott, E., Saupe, T., Thompson, J. E., Robb, J. E., & Scheib, C. L. (2022). Sex bias in N eolithic megalithic burials. American Journal of Biological Anthropology. https://doi.org/10.1002/ajpa.24645
Guellil, M., Keller, M., Dittmar, J. M., Inskip, S. A., Cessford, C., Solnik, A., Kivisild, T., Metspalu, M., Robb, J. E., & Scheib, C. L. (2022). An invasive Haemophilus influenzae serotype b infection in an Anglo-Saxon plague victim. Genome Biology, 23(1), 1–27.
Guellil, M., van Dorp, L., Inskip, S. A., Dittmar, J. M., Saag, L., Tambets, K., Hui, R., Rose, A., D’Atanasio, E., Kriiska, A., Varul, L., Koekkelkoren, A. M. H. C., Goldina, R. D., Cessford, C., Solnik, A., Metspalu, M., Krause, J., Herbig, A., Robb, J. E., … Scheib, C. L. (2022). Ancient herpes simplex 1 genomes reveal recent viral structure in Eurasia. Science Advances, 8(30), eabo4435.
Hui, R., D’Atanasio, E., Cassidy, L. M., Scheib, C. L., & Kivisild, T. (2020). Evaluating genotype imputation pipeline for ultra-low coverage ancient genomes. Scientific Reports, 10(1), 1–8.
Hui, R., Scheib, C. L., D’Atanasio, E., Inskip, S. A., Cessford, C., Biagini, S. A., Wohns, A. W., Ali, M. Q. A., Griffith, S. J., Solnik, A., Niinemäe, H., Ge, X. J., Rose, A. K., Beneker, O., O’Connell, T. C., Robb, J. E., & Kivisild, T. (2024). Genetic history of Cambridgeshire before and after the Black Death. Science Advances, 10(3), eadi5903.
Keller, M., Spyrou, M. A., Scheib, C. L., Neumann, G. U., Kröpelin, A., Haas-Gebhard, B., Päffgen, B., Haberstroh, J., Ribera I Lacomba, A., Raynaud, C., Cessford, C., Durand, R., Stadler, P., Nägele, K., Bates, J. S., Trautmann, B., Inskip, S. A., Peters, J., Robb, J. E., … Krause, J. (2019). Ancient Yersinia pestis genomes from across Western Europe reveal early diversification during the First Pandemic (541-750). Proceedings of the National Academy of Sciences of the United States of America, 116(25), 12363–12372.
Pfrengle, S., Neukamm, J., Guellil, M., Keller, M., Molak, M., Avanzi, C., Kushniarevich, A., Montes, N., Neumann, G. U., Reiter, E., Tukhbatova, R. I., Berezina, N. Y., Buzhilova, A. P., Korobov, D. S., Suppersberger Hamre, S., Matos, V. M. J., Ferreira, M. T., González-Garrido, L., Wasterlain, S. N., … Scheib, C.L., Schuenemann, V. J. (2021). Mycobacterium leprae diversity and population dynamics in medieval Europe from novel ancient genomes. BMC Biology, 19(1), 1–18.
Saupe, T., Montinaro, F., Scaggion, C., Carrara, N., Kivisild, T., D’Atanasio, E., Hui, R., Solnik, A., Lebrasseur, O., Larson, G., Alessandri, L., Arienzo, I., De Angelis, F., Rolfo, M. F., Skeates, R., Silvestri, L., Beckett, J., Talamo, S., Dolfini, A., … Scheib, C. L. (2021). Ancient genomes reveal structural shifts after the arrival of Steppe-related ancestry in the Italian Peninsula. Current Biology, 31(12), 2576-2591.e12.
Scheib, C. L. (2021). Ancient Health Landscape: foundations and perspectives. Journal of Anthropological Sciences = Rivista Di Antropologia: JASS / Istituto Italiano Di Antropologia, 99. https://doi.org/10.4436/JASS.99011
Scheib, C. L., Hui, R., D’Atanasio, E., Wohns, A. W., Inskip, S. A., Rose, A., Cessford, C., O’Connell, T. C., Robb, J. E., Evans, C., Patten, R., & Kivisild, T. (2019). East Anglian early Neolithic monument burial linked to contemporary Megaliths. Annals of Human Biology, 46(2), 145–149.
Scheib, C. L., Hui, R., Rose, A. K., D’Atanasio, E., Inskip, S. A., Dittmar, J., Cessford, C., Griffith, S. J., Solnik, A., Wiseman, R., Neil, B., Biers, T., Harknett, S.-J., Sasso, S., Biagini, S. A., Runfeldt, G., Duhig, C., Evans, C., Metspalu, M., … Kivisild, T. (2024). Low genetic impact of the Roman occupation of Britain in rural communities. Molecular Biology and Evolution, 41(9), msae168.
Inskip, S., Scheib, C. L., Wohns, A. W., Ge, X., Kivisild, T., & Robb, J. (2019). Evaluating macroscopic sex estimation methods using genetically sexed archaeological material: The medieval skeletal collection from St John’s Divinity School, Cambridge. In American Journal of Physical Anthropology (Vol. 168, Issue 2, pp. 340–351). https://doi.org/10.1002/ajpa.23753
Scheib, C. L., Li, H., Desai, T., Link, V., Kendall, C., Dewar, G., Griffith, P. W., Mörseburg, A., Johnson, J. R., Potter, A., Kerr, S. L., Endicott, P., Lindo, J., Haber, M., Xue, Y., Tyler-Smith, C., Sandhu, M. S., Lorenz, J. G., Randall, T. D., … Kivisild, T. (2018). Ancient human parallel lineages within North America contributed to a coastal expansion. Science, 360(6392), 1024–1027.
Spyrou, M. A., Keller, M., Tukhbatova, R. I., Scheib, C. L., Nelson, E. A., Valtueña, A. A., Neumann, G. U., Walker, D., Alterauge, A., Carty, N., Cessford, C., Fetz, H., Gourvennec, M., Hartle, R., Henderson, M., von Heyking, K., Inskip, S. A., Kacki, S., Key, F. M., … Krause, J. (2019). Phylogeography of the second plague pandemic revealed through analysis of historical Yersinia pestis genomes. In Nature Communications (Vol. 10, Issue 1). https://doi.org/10.1038/s41467-019-12154-0
