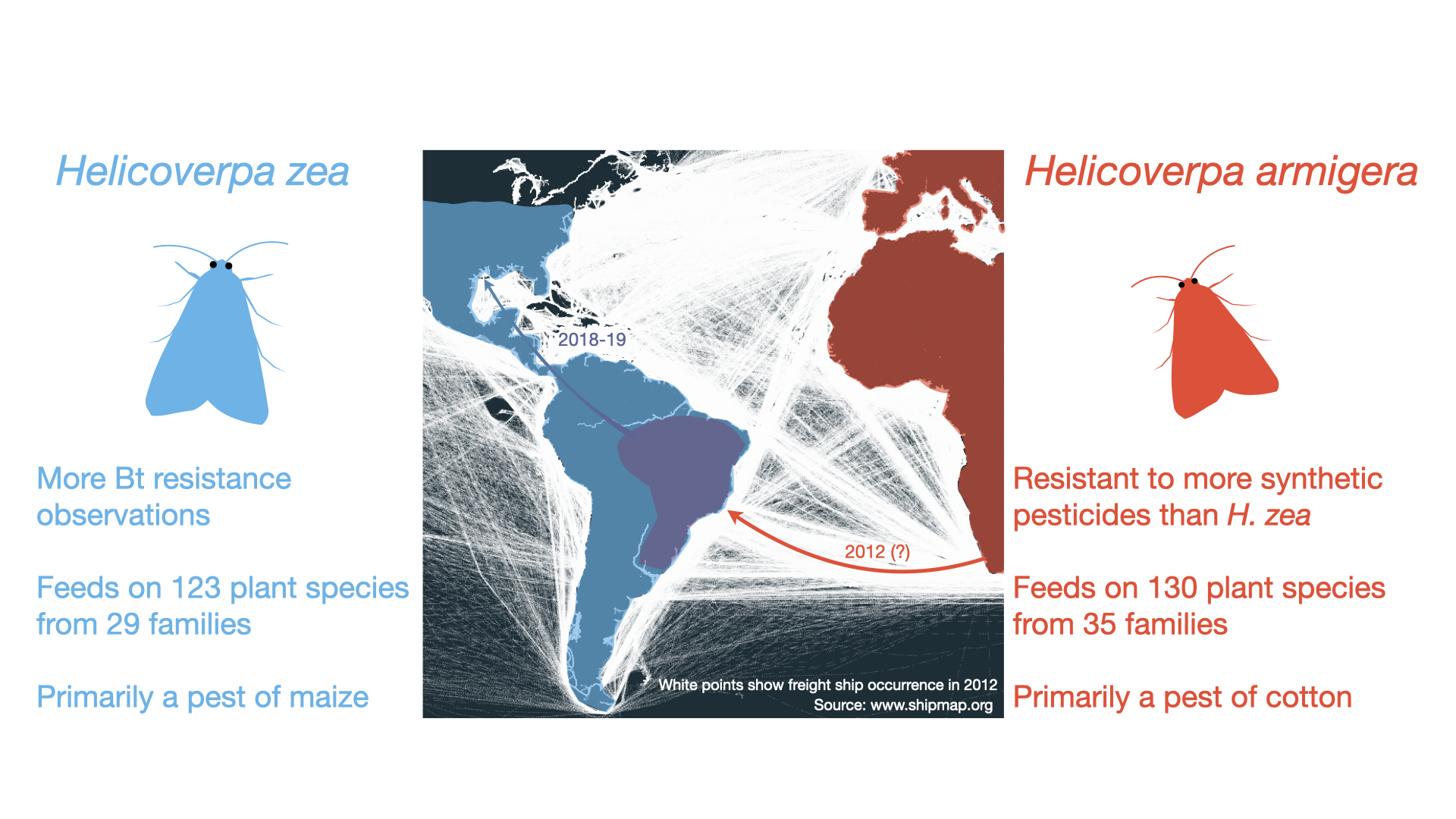
Image: Spread of Helicoverpa armigera (red) into South America. The native range of H. zea is shown in blue. The individuals from the admixed population (purple) have spread into North America. White points in the central panel show freight ship occurrence in 2012. Left: H. zea photographed in the US by Jack Cochran (top) and Rodney Haynes (bottom). Right: H. armigera photographed in Spain by Luis Orlando (top) and in Czechia by Martin Vašina (bottom). Photographs licensed from the Global Biodiversity Information Facility under http://creativecommons.org/licenses/by-nc-nd/4.0/
Population genomics of invasive Helicoverpa crop pests
Supervisor: Professor Chris Jiggins
Co-supervisor: Henry North
Noctuid moths in the genus Helicoverpa include some of the most economically damaging agricultural pests in the world. H. armigera is particularly damaging: this species has rapidly evolved resistance to a wide variety of pesticides, and has adapted to highly variable climatic conditions throughout its broad Afro-Eurasian native range. In 2012, H. armigera was detected as an invasive species in Brazil, where it caused crop losses estimated to be in the order of billions of US dollars per year. As the invasive population spread throughout South America, H. armigera came into secondary contact with its sister species, H. zea — also a significant crop pest, native throughout the Americas. The two species occasionally hybridized in Brazil. This resulted in the spread of a pesticide resistance allele from invasive H. armigera to native H. zea early during the biological invasion. Over time, alleles conferring a different form of resistance were introduced to invasive H. armigera from native H. zea. Despite the bi-directional exchange of fitness-enhancing alleles, the two species have remained distinct in sympatry. More recently, H. armigera has been directly introduced to North America, increasing the risk of further spread and the spillover of non-native alleles into the native H. zea population.
This system presents an extraordinary opportunity to study gene exchange across semi-permeable species boundaries and to develop a genomic monitoring strategy for tracing, and potentially forecasting, invasive spread. Extensive genomic resources have been generated, including a 10-year time series of ~1000 whole-genome sequences sampled across multiple locations in Brazil. Multiple high quality reference genomes are available for H. armigera, H. zea, and closely related species. Depending on the student’s interests, topics of enquiry could include:
• The development of a graph-based pangenome for the Helicoverpa genus
• The investigation of gene flow among other globally distributed Helicoverpa species
• The design of a low-cost amplicon panel for species and resistance diagnosis
• The population-level analysis of non-target co-bionts in genomic datasets; macro-evolutionary analysis of gene family expansions in the “megapest” lineage of heliothine moths; spatial modelling of invasive spread; and
• Investigations into the genetic and mechanistic basis of reproductive isolation between species.
Type of work
This project could involve computational, field work and laboratory experiments depending on interests of the student. Appropriate bioinformatics training will be provided, including courses from the University’s Bioinformatics and Statistics Training programmes. This will require an interest in, and some familiarity with, coding in languages such as Python, Unix/Bash, and R. A familiarity with population genetics and bioinformatics will be advantageous. The student will carry out population genomic analyses using the University of Cambridge Clinical School Computing Cluster. These analyses will include detecting evidence of selection, quantifying gene flow within and between species, performing gene-environment association tests, and inferring demographic history. The student will generate or utilize existing pangenome graphs, with opportunities to develop novel graph-based methods for analysis and visualisation. The specific analyses will depend on the interests of the student, and will be developed in collaboration with lead and co-supervisors. New genomic datasets could potentially be generated over the course of the PhD, though this is not essential to the aims of the project.
Importance of the area of research concerned
Invasive species pose a significant and growing threat to global food security, biodiversity, and human health. Many invasive insect species are agricultural pests, currently destroying 5-20% of all grain crop production. The extent of agro-economic damage is expected to increase because of climate change, the continued spread of invasive species, and the evolution of pesticide resistance. Population and evolutionary genomics provide powerful tools for monitoring, managing, and mitigating the spread of invasive species and pesticide resistance. At the same time, invasive insect pests represent remarkable and otherwise-impossible ‘experiments in evolution’, which can be used to investigate evolutionary dynamics in ‘real time’. This project is an opportunity to apply perspectives and analytical techniques developed in the field of evolutionary biology to one of the most pressing challenges of the Anthropocene.
References
Anderson, Craig J., et al. Hybridization and gene flow in the mega-pest lineage of moth, Helicoverpa. Proceedings of the National Academy of Sciences 115.19 (2018): 5034-5039.
Valencia-Montoya, Wendy A., et al. Adaptive introgression across semipermeable species boundaries between local Helicoverpa zea and invasive Helicoverpa armigera moths. Molecular biology and evolution 37.9 (2020): 2568-2583.
North, Henry L., et al. Rapid adaptation and interspecific introgression in the North American crop pest Helicoverpa zea. Molecular Biology and Evolution 41.7 (2024): msae129.
Jin, Minghui, et al. Adaptive evolution to the natural and anthropogenic environment in a global invasive crop pest, the cotton bollworm. The Innovation 4.4 (2023).
North, Henry L., Angela McGaughran, and Chris D. Jiggins. Insights into invasive species from whole‐genome resequencing. Molecular Ecology 30.23 (2021): 6289-6308.
McGaughran, Angela, et al. Genomic tools in biological invasions: Current state and future Frontiers. Genome biology and evolution 16.1 (2024): evad230.
Ålund, Murielle, et al. Anthropogenic Change and the Process of Speciation. Cold Spring Harbor Perspectives in Biology 15.12 (2023): a041455.
