I am passionate about the evolution of morphological diversity. What are the genes, mutations and developmental mechanisms responsible for the emergence of morphological variation? To answer this question we combine developmental genetics, quantitative and population genetics approaches using Lake Malawi cichlid fishes as a model system.
Mechanisms underlying the emergence and diversification of pigmentation patterns: The pigmentation patterns of lake Malawi cichlid fishes provide an ideal system to study morphological evolution. We specifically focus on a set of brightly pigmented spots on male anal fins, known as “egg-spots”. Egg-spots are a diverse trait (number, colour and shape), and play a key role in the territorial and breeding behaviour of around 1,500 species of cichlids. They are relatively simple structure, consisting of a circular area made up of xanthophores and iridophores (orange and silver pigment cells respectively) and are often surrounded by a transparent outer-ring. With the availability of hundreds of cichlid species genomes and genome editing tools (eg. Tol2 transgenesis and CRISPR), we can link genetics, cellular biology, development, to organismal trait variation. By addressing these mechanisms across different timescales (populations, species and genera) we can also tackle a long-standing question: whether the basis of adaptive change is the same at the micro- and macro-evolutionary scale.
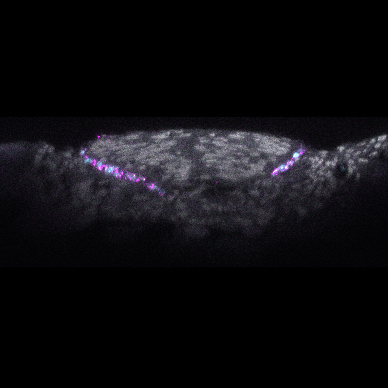
Neural crest cell development and evolution in cichlid fishes:
Neural crest cells are a multipotent embryonic cell population that emerge from the vertebrate dorsal neural tube during early development. They then delaminate and undergo some of the longest migrations of any embryonic cell type to give origin to multiple derivatives such as pigment cells, neurons and glia of the peripheral nervous system, smooth muscle, craniofacial cartilage and bone. Many of the features that distinguish vertebrates from their chordate relatives are derived from the neural crest, making this multipotent embryonic cell population a key innovation central to vertebrate evolution. The origin and development of the neural crest has been extensively studied in a handful of model organisms (e.g. lamprey, zebrafish, frog and chicken). Most work focusing on the gene regulatory networks and developmental mechanisms associated with NC specification and differentiation, together with the examination of how one stem cell population can migrate so extensively to different parts of the vertebrate body plan and diversify into multiple cell types. However, we still do not know how variation in NC genetic and developmental mechanisms underlie the subtle variation in NC derived morphologies (e.g. craniofacial skeleton and pigmentation) present within a population and between closely related species. We are currently addressing this using cichlid natural variation to identify genes and cellular mechanisms generating variation during NC development.